10: Biohydrogen - Hydrogenases
- Page ID
- 34465
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Search Fundamentals of Biochemistry
-
Conceptualize Hydrogen as a Clean Fuel:
- Explain the oxidation reaction of H₂ and why its combustion produces only water, minimizing greenhouse gas emissions.
- Compare the energy density of hydrogen with other common fuels.
-
Differentiate Production Methods and “Hydrogen Colors”:
- Describe the various production methods for hydrogen (biological, electrochemical/electrolytic, and thermochemical).
- Understand the classification of hydrogen by “color” (green, blue, grey, black, etc.) based on environmental impact and production process.
-
Understand Biological H₂ Production Mechanisms:
- Explain how hydrogenases catalyze the reversible conversion of protons and electrons to H₂.
- Distinguish between the roles of Ni–Fe and [FeFe] hydrogenases in hydrogen oxidation and production.
- Describe the general reaction schemes that illustrate H₂ serving as both an electron donor and electron sink.
-
Explore Electron Transfer and Bifurcation:
- Relate hydrogen oxidation and reduction reactions to cellular electron transport processes (e.g., mitochondrial electron transport chain and photosynthesis).
- Explain the concept of electron bifurcation in hydrogenase complexes and its significance in redox balancing.
-
Examine Industrial Implications and Biomimetic Catalysis:
- Discuss how insights from hydrogenases inspire the design of transition metal active site mimetics for industrial-scale H₂ production.
- Evaluate the challenges of scaling up biological hydrogen production compared to electrochemical and thermochemical methods.
-
Analyze Oxygen Sensitivity and Protective Strategies:
- Understand why many hydrogenases are oxygen-sensitive and how this impacts their practical application.
- Describe strategies (e.g., use of CO or sulfides) that protect hydrogenase active sites from oxygen-induced damage.
These goals aim to integrate knowledge of biochemical redox processes, enzyme mechanisms, and emerging sustainable energy technologies, providing a foundation for further study and application in the field of bioenergy.
Introduction
In the last section, we described different ways to produce H2 and the colors ascribed to them based on the environmental impacts. Many look to produce and use H2 to provide energy without releasing CO2. As shown in the reaction below, H2 can be used in fuel cells to power spacecraft and cars.
\begin{equation}
\begin{aligned}
& \mathrm{O}_2+4 \mathrm{H}^{+}+4 \mathrm{e}^{-} \longrightarrow 2 \mathrm{H}_2 \mathrm{O} \\
& \mathrm{H}_2 \longrightarrow 4 \mathrm{H}^{+}+4 \mathrm{e}^{-}
\end{aligned}
\end{equation}
Cars that run on H2, considered a zero-emission fuel, are already available. These fully electric cars use fuel cells powered by the oxidation of H2 to produce electrical energy.
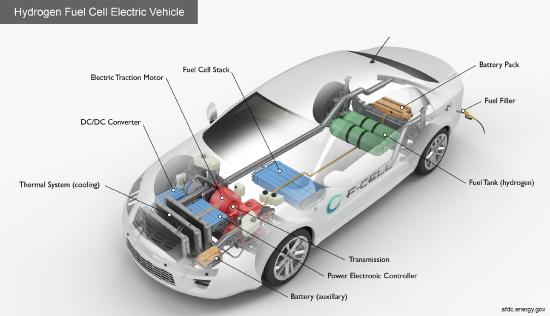
Figure \(\PageIndex{1}\): https://afdc.energy.gov/vehicles/how...tric-cars-work
Given the scale needed, most H2 is derived from the steam reformation of natural gas and water electrolysis. From a biochemical perspective, cells have evolved to make H2 and use H2 as an energy source. Direct microbial production of H2 is unlikely to meet society's energy needs. The 2023 International Energy Agency (IEA) report, "Hydrogen patents for a clean energy future", doesn't mention direct microbial production. However, much can be learned by studying how hydrogenases (H2ases, Hyd), enzymes that make or use H2, work. (Don't confuse hydrogenases with dehydrogenases that directly use NAD+/NADH and FAD/FADH2) for redox reactions. Transition metal active site mimetics can be made as potential catalysts for more industrial-level production of H2.
Although the reversible formation of H2 involves the most elemental particles in chemistry, protons (H+) and electrons (e-), the biological reactions that produce and consume H2 are complex. Before we proceed, let's see how these reactions are similar to other biochemical reactions and pathways we have already discussed.
Use of H2 as a source of electrons for reduction reactions.
Each hydrogen in H2 has an oxidation number of 0. Each can be oxidized to H+ (oxidation number +1) with the 2 electrons passed on to a substrate/cofactor or a sequential series of substrates with higher and higher standard reduction potentials (better oxidizing agents), forming reduced products.
H2 + (substrate)OX → 2H+ + (product)RED
This general reaction is analogous to the mitochondrial electron transport chain, in which electrons are passed from a source (NADH) to oxidized forms of acceptors. The general reaction below shows each redox pair in the electron transport chain.
NADH/NAD+ → FAD/FADH2 → UQ/UQH2 → Cyto COX/Cyto CRED → O2/H2O
Some organisms have evolved to produce energy by oxidizing H2, which is analogous to the photosynthetic oxidation of water. In photosystem II, the oxygen-evolving complex oxidizes oxygen in H2O (oxidation number -2) as it forms O2 (oxidation number 0). Some redox pairs, starting with H2O/O2, are shown below for photosystem II.
H2O/O2 → P680/P680* → (Plastoquione)OX/(Plastoquione)RED
The first reaction is endergonic and requires photons as an energy source.
Use of H+ as a sink for electrons for oxidation reactions that produce H2.
H+ has an oxidation number of +1. Hence, it can be reduced to H2 (oxidation number of 0) as it gains electrons from substrates/cofactors, which get oxidized. This general reaction is shown below.
2H+ + (substrate/cofactor)RED → H2 + (substrate/cofactor)OX
Many microorganisms can produce H2 through variants of photosynthesis or fermentation, both of which provide the two electrons needed. E. coli has four hydrogenases (Hyd 1, 2, 3, and 4). It forms H2 through two reactions catalyzed by:
- formate (HCO2-) dehydrogenase (FDH): 2HCO2- ⇌ 2CO2 + 2H+ + 2e-
- hydrogenase (H2ase): 2H+ + 2e- → H2
The C in formate has an oxidation number of +2 and is oxidized to CO2, in which the C has an oxidation number of +4.
The formate hydrogenlyase (FHL) complex contains both the formate dehydrogenase (FDH) and a hydrogenase (H2ase) and reversibly interconverts HCO2– and H2. The E. coli FHL-1 complex, which makes H2 using fermentation, is shown below in Figure \(\PageIndex{2}\). The complex can be immobilized on a Macro-mesoporous inverse opal (IO) indium tin oxide (ITO) electrode (IO-IPO) or ITO nanoparticles (NP), which can relay electrons.
Figure \(\PageIndex{2}\): Katarzyna P. Sokol et al. J. Am. Chem. Soc. 2019, 141, 44, 17498–17502. https://doi.org/10.1021/jacs.9b09575. CC-BY license
Panel (a) shows the biological E. coli FHL-1 complex. FdhF, [Mo]-FDH; B/F/G, Fe–S cluster-containing proteins; HycE, [NiFe]-H2ase; HycD/C, membrane proteins. (17)
Panel (b) shows a IO-ITO|FDH||IO-ITO|H2ase cell: IO-ITO|FDH wired to IO-ITO|H2ase electrode.
Panel (c) shows a FDH–ITO–H2ase nanoparticle (NP) system with enzymes immobilized onto ITO NP in solution. Species size not drawn to scale.
All you need to synthesize H2 are 2 protons and 2 electrons (potentially derived from photosynthesis). Let's take a deeper look at the hydrogenase that catalyzes H2 production.
Hydrogenases (H2ases)
Hydrogenases catalyze the reversible conversion of 2H+ → H2. A hydrogenase database, HydDB, a web tool for hydrogenase classification and sequence analysis, shows their high diversity and metabolic roles. There are three classes of hydrogenases: the Ni-Fe (most abundant, primarily for H2 conversion to 2H+), the Fe-Fe (highest kcat for H2 production), and the single Fe hydrogenases, as shown in Table \(\PageIndex{2}\) below. We won't discuss the single Fe hydrogenases.
| CLASSES AND SUBCLASSES OF HYDROGENASES | ||
| [NiFe] Group 1: Respiratory H2-uptake [NiFe]-hydrogenases | ||
| 1a | Periplasmic | Electron input for sulfate, metal, and organohalide respiration. [NiFeSe] variants. |
| 1b | Prototypical | Electron input for sulfate, fumarate, metal, and nitrate respiration. |
| 1c | Hyb-type | Electron input for fumarate, nitrat,e and sulfate respiration. Physiologically reversible. |
| 1d | Oxygen-tolerant | Electron input for aerobic respiration and oxygen-tolerant anaerobic respiration. |
| 1e | Isp-type | Electron input primarily for sulfur respiration. Physiologically reversible. |
| 1f | Oxygen-protecting | Unresolved role. May liberate electrons to reduce reactive oxygen species. |
| 1g | Crenarchaeota-type | Electron input primarily for sulfur respiration. |
| 1h | Actinobacteria-type | Electron input for aerobic respiration. Scavenges electrons from atmospheric H2. |
| 1i | Coriobacteria-type (putative) | Undetermined role. May liberate electrons for anaerobic respiration. |
| 1j | Archaeoglobin-type | Electron input for sulfate respirationπ. |
| 1k | Methanophenazine-reducing | Electron input for methanogenic heterodisulfide respiration. |
| [NiFe] Group 2: Alternative and sensory uptake [NiFe]-hydrogenases | ||
| 2a | Cyanobacteria-type | Electron input for aerobic respiration. Recycles H2 produced by other cellular processes. |
| 2b | Histidine kinase-linked | H2 sensing. Activates a two-component system controlling hydrogenase expression. |
| 2c | Diguanylate cyclase-linked (putative) | Undetermined role. May sense H2 and regulate processes through cyclic di-GMP production. |
| 2d | Aquificae-type | Unresolved role. May generate reductant for carbon fixation or have a regulatory role. |
| 2e | Metallosphaera-type (putative) | Undetermined role. May liberate electrons primarily for aerobic respiration. |
| [NiFe] Group 3: Cofactor-coupled bidirectional [NiFe]-hydrogenases | ||
| 3a | F420-coupled | Couples oxidation of H2 to the reduction of F420 during methanogenesis. Physiologically reversible. [NiFeSe] variants. |
| 3b | NADP-coupled | Couples oxidation of NADPH to the evolution of H2. Physiologically reversible. May have sulfhydrogenase activity. |
| 3c | Heterodisulfide reductase-linked | Bifurcates electrons from H2 to heterodisulfide and Fdox in methanogens. [NiFeSe] variants. |
| 3d | NAD-coupled | Interconverts electrons between H2 and NAD depending on cellular redox state. |
| [NiFe] Group 4: Respiratory H2-evolving [NiFe]-hydrogenases | ||
| 4a | Formate hydrogenlyase | Couples formate oxidation to fermentative H2 evolution. May be H+-translocating. |
| 4b | Formate-respiring | Respires formate or carbon monoxide using H+ as an electron acceptor. Na+-translocating via Mrp. |
| 4c | Carbon monoxide-respiring | Respires carbon monoxide using H+ as an electron acceptor. H+-translocating. |
| 4d | Ferredoxin-coupled, Mrp-linked | Couples Fdred oxidation to H+ reduction. Na+-translocating via Mrp complex. |
| 4e | Ferredoxin-coupled, Ech-type | Couples Fdred oxidation to H+ reduction. Physiologically reversible via H+/Na+ translocation. |
| 4f | Formate-coupled (putative) | Undetermined role. May couple formate oxidation to H2 evolution and H+ translocation. |
| 4g | Ferredoxin-coupled (putative) | Undetermined role. May couple Fdred oxidation to proton reduction and H+/Na+ translocation. |
| 4h | Ferredoxin-coupled, Eha-type | Couples Fdred oxidation to H+ reduction in anaplerotic processes. H+/Na+-translocating. |
| 4i | Ferredoxin-coupled, Ehb-type | Couples Fdred oxidation to H+ reduction in anabolic processes. H+/Na+-translocating. |
| [FeFe] Hydrogenases | ||
| A1 | Prototypical | Couples ferredoxin oxidation to fermentative or photobiological H2 evolution. |
| A2 | Glutamate synthase-linked (putative) | Undetermined role. May couple H2 oxidation to NAD reduction, generating reductant for glutamate synthase. |
| A3 | Bifurcating | Reversibly bifurcates electrons from H2 to NAD and Fdox in anaerobic bacteria. |
| A4 | Formate dehydrogenase-linked | Couples formate oxidation to H2 evolution. Some bifurcate electrons from H2 to ferredoxin and NADP. |
| B | Colonic-type (putative) | Undetermined role. May couple Fdred oxidation to fermentative H2 evolution. |
| C1 | Histidine kinase-linked (putative) | Undetermined role. May sense H2 and regulate processes via histidine kinases. |
| C2 | Chemotactic (putative) | Undetermined role. May sense H2 and regulate processes via methyl-accepting chemotaxis proteins. |
| C3 | Phosphatase-linked (putative) | Undetermined role. May sense H2 and regulate processes via serine/threonine phosphatases. |
| [Fe] Hydrogenases | ||
| All | Methenyl-H4MPT dehydrogenase | Reversibly couples H2 oxidation to 5,10-methenyltetrahydromethanopterin reduction. |
Dan Søndergaard et al., Scientific Reports volume 6, Article number: 34212 (2016). Creative Commons Attribution 4.0 International License. http://creativecommons.org/licenses/by/4.0/.
The three main types have different main functions in general. The Ni-Fe, Fe-Fe, and Fe H2ases generally oxidize H2, produce H2, and promote H- (hydride) transfer, respectively, as shown in Figure \(\PageIndex{3}\) below.
Figure \(\PageIndex{3}\): The active site structures of [NiFe] H2ases that mainly catalyze H2 oxidation reactions, [FeFe] H2ases that mainly catalyze H2 evolution reactions, and [Fe] H2ases that catalyze H−transfer to the substrate via heterolytic H2 cleavage. X, possible H2 active sites; Y, methenyltetrahydromethanopterin; GMP, guanosine monophosphate. Seiji Ogo et al., Science Advances.(2020). DOI: 10.1126/sciadv.aaz81. Creative Commons Attribution-NonCommercial License 4.0 (CC BY-NC).
Much effort has been devoted to making transition state analogs of the active site to act as catalysts for H2 production for fuel cells. Transition metal catalysts that mimic the structures and activities of the three hydrogenases have been made. Figure \(\PageIndex{4}\) shows three specific ones below.
Figure \(\PageIndex{4}\): The differing reactivity of the three isomers. Y′, methylene blue [MB]+.Seiji Ogo et al., ibid.
The ligand containing P and PH is bis(diphenylphosphino)ethane.
First, we will explore the Ni-Fe hydrogenases.
Ni-Fe H2ases (Hyd):
We'll discuss two examples of Ni-Fe H2ases
Group 1a periplasmic (membrane-bound) hydrogenases - MBH
These are used in fuel cells and H2-producing devices since they can adhere to surfaces that can be useful, heterogeneous (not in solution) catalysts. O2 also damages some. The enzyme consists of a large subunit, found in the periplasm, and a small subunit, which anchors the protein in the plasma membrane of bacteria. This enzyme oxidizes H2: H2 → 2H++2e−. The electrons enter the bacterial respiratory chain through quinones. The transmembrane part of the small subunit binds cytochrome b, which is involved in electron transfer with the quinones, as we saw in Complex II of mitochondrial electron transport. Some soil bacteria (like Ralstonia eutropha,) can use H2 as their sole energy source. The orientation of a NiFe MBH within a bacterial cell is shown in Figure \(\PageIndex{5}\) below.
Figure \(\PageIndex{5}\): The orientation of a NiFe MBH within a bacterial cell. Lindsey A. Flanagan* and Alison Parkin. Biochem Soc Trans. 2016 Feb 15; 44(1): 315–328 (2016). doi: 10.1042/BST20150201 Creative Commons Attribution Licence 3.0.
Panel (A) shows a cartoon depiction of a NiFe MBH's location within the cytoplasmic membrane. White boxes represent the redox-active metal centers, and blue, orange, and purple blocks indicate the large, small, and cytochrome subunits, respectively.
Panel (B) shows how the E. coli hydrogenase-1 large (blue ribbon), small (orange ribbon), and cytochrome (purple ribbon) subunits can interact.
Figure \(\PageIndex{6}\) shows an interactive iCn3D modelof the O2-Tolerant Membrane-Bound Hydrogenase 1 from Escherichia coli in Complex with Its Cognate Cytochrome b (4GD3). The same color coding is used for the subunits as in the above figures.
Figure \(\PageIndex{6}\): O2-Tolerant Membrane-Bound Hydrogenase 1 from Escherichia coli in Complex with Its Cognate Cytochrome b (4GD3). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...9p4CSQRDPM8nL7
The large and small chains of hydrogenase are shown in blue and orange, respectively, while cytochrome b is shown in magenta. Cofactors and key metals are shown in spacefill. F4S is the Fe4-S3 cluster, SF4 is the iron-sulfur cluster, HEM is hemoglobin, FCO is carbonmonoxide-(dicyano) Fe, and Ni is nickel.
The biological (functional) unit consists of two heterodimers. At low O2 levels, one dimer can reactivate the other if exposed to O2. The enzyme is found in the highest concentration during anaerobic fermentation. Remember that E. coli is a facultative anaerobe that can shift its metabolic pathways to fit conditions. Perhaps its primary role is to reduce O2 to water and protect enzymes sensitive to it. Cytochrome b may mostly anchor the dimeric H2ase in the membrane.
A bifurcating Ni-Fe H2ase
These enzymes are more complicated. They oxidize H2 and move the two electrons through a complicated path that bifurcates electron flow to different substrates/cofactors. They move an electron to a low-potential (i.e., not a great oxidizing agent), high-energy species, which gets reduced in an endergonic process. The other electron simultaneously moves to a high-potential (i.e., great oxidizing agent), low-energy species that also gets reduced in an exergonic process. The overall electron transfer is thermodynamically favorable. An example might prove helpful. NADH (E0' = -280 mV, higher potential) can reduce the protein ferredoxin (E0' = -500 mV, lower potential), which can then pass its electrons in other reactions, including the formation of H2, CH4, and NH3. ATP is not required.
Four classes of bifurcating enzymes that use FAD/FADH2 or FMN/FMNH2 are known. They are optimal since they can participate in either 1 or 2 electron transfers. We will see an example of a Fe-Fe H2ase further below.
In electron transport, we encountered an electron bifurcating complex in the Q-cycle of Complex III. Reduced ubiquinone (UQH2, or ubiquinol) is oxidized, and the two lost electrons are bifurcated to cytochrome C in a high-potential pathway and to UQ to reform UQH2, as shown in Figure \(\PageIndex{7}\) below.
Figure \(\PageIndex{7}\): Electron bifurcation in Complex III
One example of a bifurcating Ni-Fe H2ase is the NiFe-HydABCSL protein from the bacteria A. mobile. The general structure of the pentameric form of the functional decamer is shown in Figure \(\PageIndex{8}\) below.
Figure \(\PageIndex{8}\): Structure of the A. mobile NiFe-HydABCSL pentamer. XIANG FENG et al. SCIENCE ADVANCES. 2022. DOI: 10.1126/sciadv.abm7546. Creative Commons Attribution License 4.0 (CC BY). https://creativecommons.org/licenses/by/4.0/
The five subunits are called HydA (Hyd = hydrogenase), HydB, HydC, HydL (large subunit), and HydS (small subunit). Pane (A) shows the domain structure of the five subunits. The NiFe-HydB NTD and CTDs are partially flexible, as indicated by dashed outlines. Panel (B) shows the subunit organization of the NiFe-Hyd complex and their associated cofactors.
NiFe-HydABCSL hydrogenase can reversibly oxidize H2 with the two electrons, reducing ferredoxin in an endergonic process and reducing NAD in an exergonic process. FMN is surrounded by a FeS cluster and appears to be the center of bifurcation. The reaction is as follows:
- The HydL oxidizes H2 with two electrons passing through the FeS centers in HydA to HybB.
- The electrons are passed to FMN, where the bound NAD gets reduced.
Figure \(\PageIndex{9}\) jdkfjdkjfdkjff
Figure \(\PageIndex{9}\): Proposed mechanism of electron bifurcation/confurcation in A. mobile NiFe-HydABCSL.
(A) Overall electron transfer pathway, highlighting the three branches of the electron transfer path. The mid-potential path is a black dashed line, the exergonic path is a blue dashed line, and the endergonic path is a red dashed line. (B) Conformational changes in the HydBC bifurcation core from the electron bifurcation state (BR state) to the electron transduction state (PB state).
Figure \(\PageIndex{10}\) shows an interactive iCn3D modelof the electron bifurcating Ni-Fe hydrogenase complex HydABCSL in FMN/NAD(H) bound state 7T30
Figure \(\PageIndex{10}\): Electron bifurcating Ni-Fe hydrogenase complex HydABCSL in FMN-NAD(H) bound state (7T30). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...G9nKGwqvuuh6G7
Figure \(\PageIndex{11}\) shows an interactive iCn3D modelof the cofactors in the electron bifurcating Ni-Fe hydrogenase complex HydABCSL in FMN/NAD(H) bound state 7T30
Figure \(\PageIndex{11}\): Cofactors in the electron bifurcating Ni-Fe hydrogenase complex HydABCSL in FMN-NAD(H) bound state (7T30). (Copyright; author via source). Click the image for a popup, or use this external link: https://structure.ncbi.nlm.nih.gov/i...hHuzaWUDYCMnAA.
Zoom into the Ni-Fe center catalytic site. The ligands that form coordinate covalent bonds to the Fe are called FCC, or carbon monoxide-(dicyano)-Fe, Figure \(\PageIndex{12}\) below. There are also bridging sulfurs between Fe and Ni.
Figure \(\PageIndex{12}\): Carbonmonoxo-dicyano-Fe and is shown in detail in
Fe-Fe hydrogenases
These enzymes catalyze various reactions as illustrated in Figure \(\PageIndex{13}\) below.
Figure \(\PageIndex{13}\): [FeFe]-hydrogenases phylogeny and known functions. Morra S. Front Microbiol. 2022 Mar 2;13:853626. doi: 10.3389/fmicb.2022.853626. PMID: 35308355; PMCID: PMC8924675. Creative Commons Attribution License (CC BY)
As previously proposed, a phylogenetic tree shows the phylogeny of [FeFe]-hydrogenase sequences from public databases. Enzymes experimentally characterized are indicated on the tree to show their relative position. The proposed physiological function of each enzyme is also presented where known. Hyd, hydrogenase subunit; FdhF, formate dehydrogenase subunit; Fdrex/ox, reduced/oxidized ferredoxin; NADH/NAD+, reduced/oxidized nicotinamide adenine dinucleotide. They are found in prokaryotic and eukaryotic microorganisms but not in Archaea.
These are the most active for H2 production with a kcat around 10,000 s-1. They contain a Fe2S2 cluster with CO and CN ligands forming bonds to the iron, with the iron ions bridged by a -SCH2-NH-CH2S- (aza-dithiolate). A cysteine links the Fe2S2 to a Fe4S4 cluster. These two are called the H-cluster (or [Fe]H. Within this class are a soluble, monomeric cytoplasmic form, a heterodimeric periplasmic form, and a soluble, monomeric chloroplastic form. This one has a ferredoxin, connecting it to the electron transport chain in photosynthesis. Some in this group, using both NADH and ferredoxin, are called bifurcating types, as they send two electrons from a donor in two different directions. More on this later.
They contain multiple FeS clusters. The H-cluster consists of a Fe2S2 linked to a Fe4S4 cluster (cubane-like) by a cysteine. The Fe2S2 group has CO and CN ligands, with the two Fe ions of the Fe2S2 unit coordinated by an azadithiolato ligand, as shown below in Figure \(\PageIndex{14}\).
Figure \(\PageIndex{14}\): Chemical structure of the H-cluster, which is the active site of the [FeFe] hydrogenase enzyme. Rakesh C. Puthenkalathil et al., Phys. Chem. Chem. Phys., 2020, 22, 10447. https://pubs.rsc.org/fa/content/arti.../cp/c9cp06770a. This article is licensed under a Creative Commons Attribution 3.0 Unported Licence
The Fe in the [2Fe2S] cluster is linked to the cubane structure [4Fe-4S] and has six ligands, so it is saturated. The other Fe has an extra coordination site denoted by X, which can bind H+ or H2. The cluster is buried in a hydrophobic catalytic site, which helps restrict O2 access.
As we did for the Fe-Ni H2ases, we will study two examples of Fe-Fe H2ases.
Fe-Fe hydrogenase (CpI) from Clostridium pasteurianum
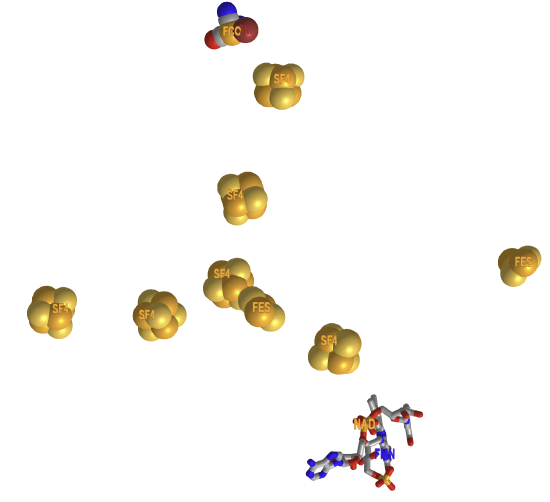
Figure \(\PageIndex{15}\) shows an interactive iCn3D modelof the H-Cluster (HC1) of Fe-Fe hydrogenase (CpI) from Clostridium pasteurianum (1FEH).
Figure \(\PageIndex{15}\): H-Cluster (HC1) of Fe-Fe hydrogenase (CpI) from Clostridium pasteurianum (1FEH). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...TksW8FGn7crh39
In this model, the CN ligands are all displayed as CO. The sulfurs are shown in green. Hover over the atoms/ions to identify them. (In iCn3D, choose, Select, Select on 3D, atom). The [4Fe-4S] subcluster forms coordinate covalent bonds with four cysteines (300, 355, 499, and 503), with one cysteine (503) forming a bridge to the [2Fe] cluster. The Fe ions in that cluster have an octahedral arrangement of ligands surrounding them. One of the ligands is water (no connecting C atom).
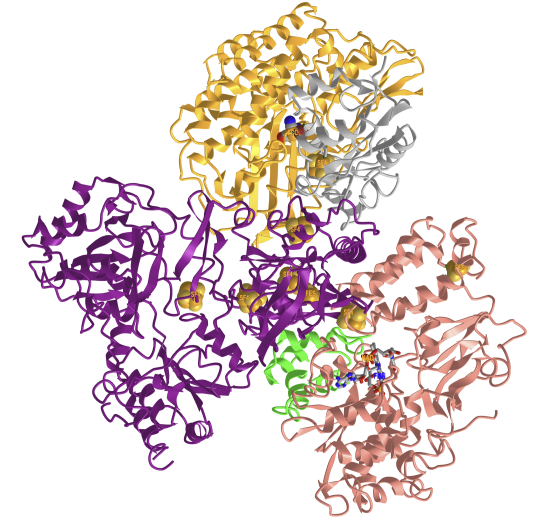
Figure \(\PageIndex{16}\) shows an interactive iCn3D modelof the Fe-Fe hydrogenase (CpI) from Clostridium pasteurianum (1FEH)
Figure \(\PageIndex{16}\): Fe-Fe hydrogenase (CpI) from Clostridium pasteurianum (1FEH). (Copyright; author via source).
Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...QJsoZ8zbvkpDc7.
As we mentioned above, the net reaction is:
2H+ + (substrate/cofactor)RED → H2 + (substrate/cofactor)OX
Many microorganisms can produce H2 through variants of photosynthesis or fermentation, both of which provide the two electrons needed. E. Coli has four hydrogenases (Hyd 1, 2, 3, and 4). It forms H2 through two reactions catalyzed by:
- formate (HCO2-) dehydrogenase: 2HCO2- ⇌ 2CO2 + 2H+ + 2e-
- hydrogenase 3: 2H+ + 2e- → H2
Figure \(\PageIndex{17}\) below shows a reaction scheme for the production of H2 linked to photosystem I in the chloroplast of microalgae under anaerobic conditions. It starts with the absorption of a photon by P700, which, in the excited state, transfers an electron to a 4Fe4S cluster in ferredoxin. This passes an electron to the HC-cluster and then onto H+.
Figure \(\PageIndex{17}\): Schematic representation of electron flow from Photosystem I to an [FeFe]-hydrogenase via a ferredoxin redox mediator (Photosystem I). JuanAmaro-Gahete et al.,Coordination Chemistry Reviews. 448, December 2021. https://doi.org/10.1016/j.ccr.2021.214172. Creative Commons CC-BY
A possible mechanism for forming H2 in the H cluster is shown below in Figure \(\PageIndex{18}\).
Figure \(\PageIndex{18}\): Proposed mechanistic cycle for hydrogen evolution in the H cluster by [FeFe]-hydrogenase adapted from Lubitz et al. JuanAmaro-Gahete et al., ibid
Start at the top left, which shows the rest of the oxidized state. In the enzyme's most oxidized resting system (Hox), the [4Fe4S] cubane is in a 2 + oxidation state while the catalytic subcluster [2Fe] is a mixed-valence FeIFeII state. The first one-electron reduction results in the formation of the Hred state, where the [4Fe4S] subcluster is reduced to a 1 + oxidation state. Protonation of the N of aza-propane-1,3-dithiolate ligand (adt-N) triggers an intramolecular charge shift to form HredH+ in which the [4Fe4S] cubane is in the 2 + state and the [2Fe] subsite is reduced to a homovalent FeIFeI state. Subsequent one-electron reduction of the subcluster [4Fe4S] gives rise to the “super-reduced” state HsredH+. In the next step of the catalytic cycle, an intermediate hydride state [Hhyd] is formed by an intramolecular proton shift from the adt-N to the distal iron Fd. This process is coupled to an electron rearrangement in the [2Fe] subsite, leading to a formal FeIIFeII oxidation state. The addition of a second proton coupled to another charge shift from the reduced [4Fe4S] to the [2Fe] subsite, either in one or two discrete steps, gives rise to [HhydH+] that is characterized by a formal FeIFeII oxidation state. At this point, there is an equilibrium between the HhydH+ and Hox[H2] in which the hydride and the proton are combined into a hydrogen molecule at the distal iron of the system. The catalytic cycle is closed by H2 release, returning to the initial Hox configuration.
A bifurcating [Fe-Fe] hydrogenase from Thermotoga maritima (HydABC)
This enzyme, functionally a heterododecamer, uses NADH as a source of electrons, which passes electrons to FMN, the bifurcation site, with an electron going to oxidized ferredoxin (Fdox) and another to H+s for reduction to FDRED and H2. The enzyme consists of a dimer of a trimer of subunits HydA, HydB, and HydC, with dimer (HydABC)2 interacting with another (HydABC)2 to form a heterododecamer, with both halves acting independently. The two trimers (HydABC) in the dimer (HydABC)2 are connected by a [4Fe–4S] cluster. A flexible loop in the B and A chain has a "closed" and "open" bridge conformation, with a nearby Zn2+ important in the loop conformation.
Figure \(\PageIndex{19}\) below shows the HydABC tetramer's cryo-EM structure and the redox cofactors' arrangement.
Figure \(\PageIndex{19}\): Cryo-EM structure of the HydABC tetramer and arrangement of the redox cofactors. Chris Furlan et al. (2022) eLife 11:e79361. https://doi.org/10.7554/eLife.79361. Creative Commons Attribution License
Panel (A) shows the unsharpened 2.3 Å map of Hyd(ABC)4 with D2 symmetry enforced, showing a tetramer of HydABC heterotrimers. All four copies of HydB and C are colored blue and green, respectively. The four HydA copies that make up the core of the complex are orange, yellow, pink, and red. The top and bottom halves of the complex are constituted by dimers of HydABC protomers (each HydABC unit is a protomer); the two protomers within the same dimer are strongly interacting, while a weaker interaction is present between the top and bottom dimers.
Panel (B) shows the HydABC dimer highlighting the iron-sulfur clusters and flavin mononucleotide (FMN) constituting the electron transfer network.
Panel (C) shows the arrangement of redox cofactors within the protein complex, with two independent, identical redox networks (dashed circles); each redox network is composed of iron-sulfur clusters belonging to a Hyd(ABC)2 unit consisting of two strongly interacting HydABC protomers.
Panel (D) shows a schematic of the electron transfer network of one of the two identical Hyd(ABC)2 units, showing edge-to-edge distances (in Å) between the various cofactors. Note that our structure is of apo-HydABC and contains only the [4Fe–4S]H subcluster of the H-cluster. The 2H+/H2 interconversion reaction in (B) illustrates the site at which this reaction occurs, but this will only occur in the fully assembled H-cluster, including [2Fe]H.
Figure \(\PageIndex{20}\) shows an interactive iCn3D modelof the electron-bifurcating [FeFe] hydrogenase from Thermotoga maritima (HydABC) (7P5H), using the same colors as the figure above.
Figure \(\PageIndex{20}\): electron-bifurcating [FeFe] hydrogenase from Thermotoga maritima (HydABC) (7P5H). (Copyright; author via source).
Click the image for a popup, or use this external link: https://structure.ncbi.nlm.nih.gov/i...2hwCUnJ9BgwJi8.
Only the dimer (HydABC)2 is shown. The A (gold), B (blue), and C (green) chains are colored as in the previous figure. The conformationally flexible loop at the C-terminal of a B chain in the closed state is shown in red. The gate also includes the C-terminal part of the A subunit near it.
Figure \(\PageIndex{21}\) shows the closed-bridge and open-bridge conformations of HydABC (the closed loop was shown in the model above).
Figure \(\PageIndex{21}\): closed-bridge and open-bridge conformations of HydABC from Thermotoga maritime.
Panel (D) shows the HydB bridge domain in the open position and its fitted model. Panel (E) shows a Zn2+ hinge region and the two possible conformations of the HydB bridge domain, open (blue) and closed (light blue).
The similarities in cofactor arrangement in the Thermotoga maritime Hyd A, B, and C subunits compared to the Nqo1, Nqo2, and Nquo3 subunits in Complex I from Thermus thermophilus (discussed in Chapter 19.1) are shown in Figure \(\PageIndex{22}\) below.
Figure \(\PageIndex{22}\): Comparison of the HydA, B and C subunits of the electron bifurcating [FeFe] hydrogenase from Thermotoga maritima with the Nqo3, 1 and 2 subunits from respiratory complex I from Thermus thermophilus.
Panel (A) shows the subunits HydA (red), HydB (blues), and HydC (green) overlaid with, respectively, Nqo3, Nqo1, and Nqo2 (all yellow) of complex I from T. thermophilus (Gutiérrez-Fernández et al., 2020, PDB: 6ZIY).
Panel (B) compares the NADH-binding site of the Nqo1 subunit of complex I from T. thermophilus (light blue) with the flavin mononucleotide (FMN) site in HydB; the high similarity suggests that NADH binds in the proximity of FMN in HydABC, similar to complex I.
Panel (C) shows an electron transfer network in HydABC compared to complex I from T. thermophilus, with edge-to-edge distances indicated in bold. The red, blue, and green dotted lines indicate the cofactors present in the HydA (Nqo3), HydB (Nqo1), and HydC (Nqo2) subunits, respectively. Note that our structure is of the apo-HydABC and lacks the [2Fe]H subcluster of the H-cluster. The 2H+/H2 interconversion reaction in (C) illustrates the site at which this reaction occurs, but this will only happen in the fully assembled H-cluster, including [2Fe]H.
Here is a link to a video showing the conformational change observed between the ‘Bridge closed forward’ (7P8N) and ‘Open bridge’ (7PN2) classes.
In the video, the HydB C-terminal iron-sulfur cluster domain is colored blue, and the HydA C-terminal iron-sulfur cluster domain is colored orange. The zinc ion (gray sphere) and ligating residues (three cysteine ligands and one histidine) are also shown. The location of the HydA C-terminal domain when the bridge is open is unknown, so it is shown transparently in both states for reference.
The geometric separation of catalytic sites and the bifurcation mechanism prevent these thermodynamically favored reactions from happening
-
H2 production from ferredoxin oxidation (in the absence of NADH oxidation)
-
NAD+ reduction by H2 (in the absence of ferredoxin reduction)
-
ferredoxin oxidation by NAD+
Oxygen Sensitivity of Fe-Fe H2ases
We have alluded that Fe-Fe H2ases can be sensitive to O2. A possible mechanism involves interacting one Fe ion (Fed, the distal Fe) with oxygen, forming damaging free radicals. As CO binds more strongly than O2 to the iron in hemoglobin, its interaction with the H-center can help protect the H2ases. Sulfides can also afford protection. These mechanisms are illustrated in Figure \(\PageIndex{23}\):
Figure \(\PageIndex{23}\): Oxygen tolerance strategies in [FeFe]-hydrogenases. Morra S, ibid.
Schematic representation of the H-cluster in the oxidized active state Hox (center). In the absence of any exogenous protectant, numerous [FeFe]-hydrogenases undergo irreversible inactivation due to H-cluster damage with loss of Fe atoms (red pathway); carbon monoxide acts as a protective agent due to its ability to form Hox-CO by binding reversibly to the H-cluster at the same site as O2 (purple pathway); in DdH, a similar mechanism occurs when sulfide binds to the H-cluster forming Hinact, via the Htrans intermediate (orange pathway); in CbA5H, a conformational change in the protein structure allows for a conserved cysteine to directly bind to the H-cluster, forming Hinact (green pathway). Fep, proximal iron atom; Fed, distal iron atom; Cys, cysteine residue.
Summary
This chapter explores hydrogen as an ideal clean fuel and examines its production and utilization from both industrial and biological perspectives. It begins by highlighting the advantages of hydrogen fuel—its combustion reaction produces only water, its exceptionally high energy content per gram, and its potential for zero greenhouse gas emissions when used in fuel cells. The discussion then shifts to the various methods of hydrogen production, which are categorized by “color” (green, blue, grey, etc.) based on their environmental impact and source materials.
A significant portion of the chapter is dedicated to the biochemical processes that generate and utilize hydrogen. It explains how enzymes known as hydrogenases catalyze the reversible conversion between protons (H⁺) and molecular hydrogen (H₂). The chapter details the roles of different hydrogenase classes—[NiFe] and [FeFe]—and illustrates their mechanisms of electron transfer and bifurcation, drawing parallels with cellular processes such as the mitochondrial electron transport chain and photosynthetic water oxidation.
The text further delves into the challenges posed by oxygen sensitivity in hydrogenases and describes various protective strategies, including the use of carbon monoxide or sulfide binding, which help preserve enzyme activity in oxygenated environments. Finally, the chapter touches on the potential of using insights from natural hydrogenase systems to develop biomimetic catalysts for industrial hydrogen production, underscoring the importance of integrating biochemical understanding with sustainable energy technology.
Overall, the chapter provides a comprehensive review of hydrogen's role as a clean fuel, the enzymatic mechanisms underlying its production in nature, and the implications for designing next-generation energy solutions.


.png?revision=1&size=bestfit&width=434&height=435)
_from_Clostridium_pasteurianum_(1FEH).png?revision=1&size=bestfit&width=322&height=357)
_from_Clostridium_pasteurianum.png?revision=1&size=bestfit&width=348&height=466)
_(7P5H).png?revision=1&size=bestfit&width=590&height=422)